Introduction
Traditional cancer diagnostics rely heavily on histological analysis to make a diagnosis. The histological process begins with a biopsy to collect tissue samples that are prepared, sliced into thin sections, and stained with Hematoxylin and Eosin (H&E) to enhance visibility. These stained tissue samples are then traditionally inspected and annotated manually by pathologists. However, this method is time-consuming, requires specialized expertise, and is subject to human variability [1].
With advancements in digital technology, the digitization of these histological slides into high-resolution whole-slide images (WSIs) has paved the way for automating tasks that are too time-consuming for histopathologists. With that, various artificial intelligence (AI) solutions have been proposed for digital pathology applications. These include automated image analysis systems that extract essential diagnostic information from WSIs, such as tumor classification, subclassification, and grading, improving pathology productivity, accuracy, and reproducibility. However, deep learning models often struggle to perform consistently when trained on entire whole-slide images due to various histological variances and inconsistencies, such as tissue folds, air bubbles, tears, focus blur, and excessive color pigment deposits, which are often found on WSIs.
To address these challenges, methods are employed to detect and exclude diagnostically irrelevant areas, allowing the focus to shift to regions of interest (ROIs) within the slide. By concentrating on these ROIs, the complexity of the analysis is reduced, leading to faster and more accurate deep learning models for automated pathology [2].
Spotlight: WSI Region of Interest Extraction Workflow
In this post, we present a workflow for ROI extraction analysis, which is based on segmentation analysis—a crucial technique for identifying and distinguishing tumor tissue from normal tissue. By combining this approach with additional image processing techniques, our workflow serves as a preliminary step for downstream tasks such as classification and tumor burden estimation.
WSI Region of Interest Extraction workflow was developed by VELSERA bioinformaticians using state-of-the-art, publicly available software for image processing and segmentation [3] [4], [5]. These tools were adapted and combined into a versatile, functional workflow optimized to leverage the computing capabilities of the CAVATICA, CGC, and BDC platforms.
The WSI Region of Interest Extraction workflow is used for processing whole slide images by segmenting tumor tissue and generating sets of patches that predominantly represent either tumor or normal tissue, as illustrated in Figure 1. The workflow supports liver, colon, and breast tissue types. Apart from segmentation and patch extraction, this workflow also supports post-processing steps like stain normalization and data augmentation, which help ensure consistency across different slides and enhance data variability for application in downstream analysis. The workflow is designed with a functional, user-friendly interface (see Figure 2) that facilitates easy image import and allows for intuitive manipulation of parameters that influence segmentation quality, the size and format of output patches, as well as options for post-processing steps. Additionally, it provides a visual report that offers insights into the segmentation analysis, helping users to better understand the results.

Figure 1: Overview of the ROI Extraction Analysis
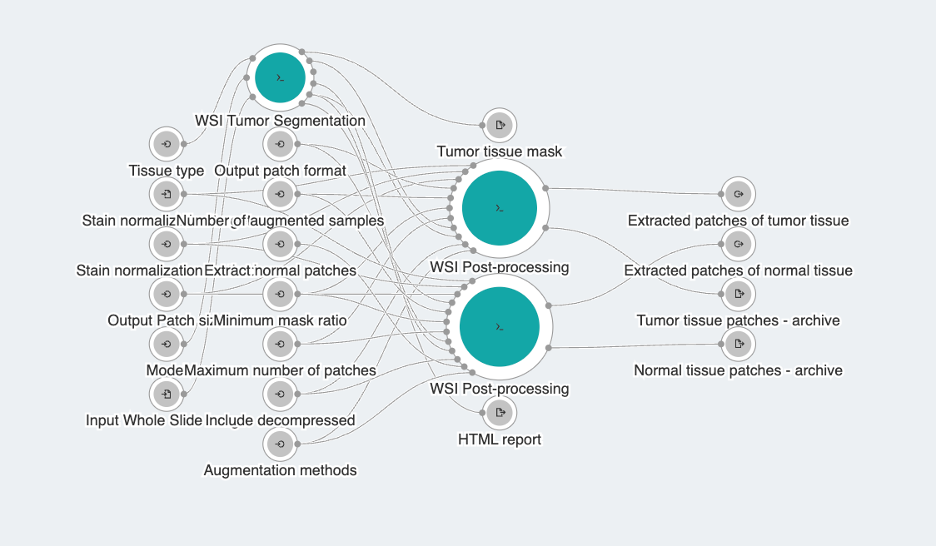
Figure 2: WSI Region of Interest Extraction workflow
Tools Overview
At the core of this workflow is the WSI Tumor Segmentation tool, complemented with utility tool for whole slide image post-processing. The workflow supports input file formats for WSI images compatible with the OpenSlide library, and for any other formats, a standalone utility tool is included in the bundle to facilitate cross-conversion of medical images. These utility tools can also be utilized independently. Below, we provide a detailed explanation of each tool’s usage and capabilities.
- The WSIC Convert tool is designed for converting Whole Slide Images (WSI) into various formats of brightfield histology images. It facilitates efficient WSI format conversion, supporting multiple input and output formats. The tool includes capabilities for reading and writing several container formats, accommodating a wide range of compression codecs, and allowing customization of tile sizes [5]. Supported input formats are: Aperio SVS (SVS), Hamamatsu (VMS, VMU, NDPI), Leica(SCN), Mirax(MRXS), Sakura(SVSLIDE), Trestle(TIF), Ventana(BIF, TIF), Generic tiled TIFF(TIF; DEFLATE, JPEG, and Webp compressed), JPEG XL, JPEG 2000, WebP, zstd, RGB/brightfield OME-TIFF, JP2 (via glymur and OpenJPG) including Omnyx JP2 files, Zarr NGFF v0.4, and DICOM VL Whole Slide Image IODs. Available output formats are: TIFF for Tiled TIFF, SVS for Aperio SVS, ZARR for Zarr (NGFF v0.4), JP2 for JPEG2000, and DCM for DICOM.
- WSI Tumor Segmentation is an advanced tool designed for the automated analysis of whole slide images, focusing on the segmentation of tumor and normal tissue regions in liver, breast, and colon tissue slides. The segmentation process involves several key steps. First, the tool identifies and isolates tissue regions from the WSI, eliminating non-tissue areas to avoid unnecessary computations. It then defines patches of tissue, which are classified as either tumor or normal using pretrained models based on an ensemble of fully convolutional networks (FCNs) [3]. These models were trained on balanced datasets containing equal numbers of tumor and normal patches, classifying a patch as tumor if even a single pixel within it is identified as such. Finally, the individual patch outputs are stitched together to generate a comprehensive segmentation of the entire WSI image. Parameters such as patch size and stride size—which defines the overlap between neighboring patches—can affect the quality of the generated tumor tissue heatmaps. These settings can be fine-tuned to achieve an optimal balance between accuracy and computational efficiency.
The WSI Tumor Segmentation tool generates NumPy array outputs for the original image, tumor tissue mask, and normal tissue mask, allowing for easier downstream manipulation. Additionally, it provides an HTML report that offers visual insights into the analysis, including images of the original WSI, the tumor tissue mask, and their overlap, along with frequently used details about the input WSI slide. - WSI Post-processing is a comprehensive tool for tissue image analysis, providing key functionalities like image patch extraction, augmentation, and stain normalization. It supports patch extraction from whole slide images using either a sliding window method, optionally filtered with input mask, or a point-based approach. The tool also offers stain normalization methods such as Reinhard and Macenko, and allows for multiple augmentations like random shear, translation, and rotation. These features enhance the robustness and diversity of machine learning models built from processed data. Notably, all transformations are optional and can be flexibly combined to suit specific user needs. For instance, if no patch size is specified, the tool uses the full image dimensions as the patch size, enabling users to apply augmentations and/or stain normalization directly to the entire image without extracting smaller patches.
By default, the tool saves the post-processed patch(es) in PNG format, packaged within a TAR.GZ archive to efficiently handle the potentially large number of files. However, users can opt for an uncompressed output or select a different image format, such as TIFF or NPY, depending on their needs.
Conclusion
Many image processing tasks are computationally intensive, which highlights the need for efficient resource optimization. The computational capabilities of our platform can be harnessed to enable automated ROI detection and analysis, which is recognized as a common step in medical image analysis across all imaging modalities. By streamlining this process, our workflow facilitates the efficient extraction of tumor and normal tissue patches from whole slide images, along with the easy application of additional post-processing steps. As a result, from a single labeled WSI, you can produce an expanded dataset of tumor and normal tissue patches, which can then be used for downstream analyses like tumor classification, grading, and other diagnostic tasks.
References
[1] Dalí F.D. dos Santos, Paulo R. de Faria, Bruno A.N. Travençolo, Marcelo Z. do Nascimento, “Automated detection of tumor regions from oral histological whole slide images using fully convolutional neural networks”, Biomedical Signal Processing and Control, Volume 69, 2021, 102921
[2] Hossain, M.S., Shahriar, G.M., Syeed, M.M.M. et al. Region of interest (ROI) selection using vision transformer for automatic analysis using whole slide images. Sci Rep 13, 11314 (2023).
[3] Khened, M., Kori, A., Rajkumar, H. et al. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci Rep 11, 11579 (2021).
[4] Pocock, J., Graham, S., Vu, Q.D. et al. TIAToolbox as an end-to-end library for advanced tissue image analytics. Commun Med 2, 120 (2022).
[5] Johnathan Pocock, Shan E Ahmed Raza, Fayyaz Minhas, Nasir Rajpoot, WSIC: a Python package and command-line interface for fast whole slide image conversion, Bioinformatics Advances, Volume 3, Issue 1, 2023, vbad122, https://doi.org/10.1093/bioadv/vbad122
